Drug Repurposing
Drug repurposing (DR), also called drug repositioning, is defined as the process of finding new uses for existing drugs. It has become an interesting alternative to de novo drug development due to the reduction in investments, risks, and time consumption.
We can define two different approaches to DR: in vitro, and in silico drug repositioning. Computational DR strategies employ modern heterogeneous biomedical data to detect novel indications for already-approved drugs.
The DISNET project aims to build a complex multilayer network that incorporates knowledge from public sources about diseases, associated symptoms, genes, drugs, drug targets, etc. Its final goal is to generate drug repurposing hypotheses.
In this context and to show its potential as a DR tool, DISNET has been used to:
- Propose a method that evaluates whether a certain drug should or not be considered to repurpose for a given disease.
- Put forward a list of repurposable drugs for COVID-19.
These two ideas have been explored in a couple studies that are explained in the following sections. Both of them developed between December 2020 and August 2021.
The researchers that participated in these studies were:
- Principal investigator: Alejandro Rodríguez González
- Main researchers: Lucía Prieto Santamaría, Marina Díaz Uzquiano, Esther Ugarte Carro
- Other researchers: Nieves Ortiz-Roldán, Yuliana Pérez Gallardo, Ernestina Menasalvas Ruiz
Evaluating DR hypotheses
The main objective of this research was to propose a new methodology for validating DR cases through biomedical integrated data. This methodology consists of identifying significant differences between DR data and non-DR data. This way, future DR case studies could be suggested and prioritized based on whether the data associated to them is significantly different to non-DR data. To achieve this objective, we have used DISNET’s biomedical-integrated data.
To develop a complete methodology that evaluates the potential of new DR hypotheses by means of biomedical integrated data, the following stages were performed:
- A list of successful DR case studies was generated from literature evidence.
- Data pre-processing was implemented to translate DR cases to DISNET vocabularies.
- Genes, symptoms and categories were analyzed to conclude whether the data from actual DR case studies presented significant differences with non-DR data. The gene and symptom analyses sought to study the therapeutic potential of the underlying mechanisms of action that shared the original and new indications for the drug. The analysis of categories was carried out to find patterns within the disease and drug groups.
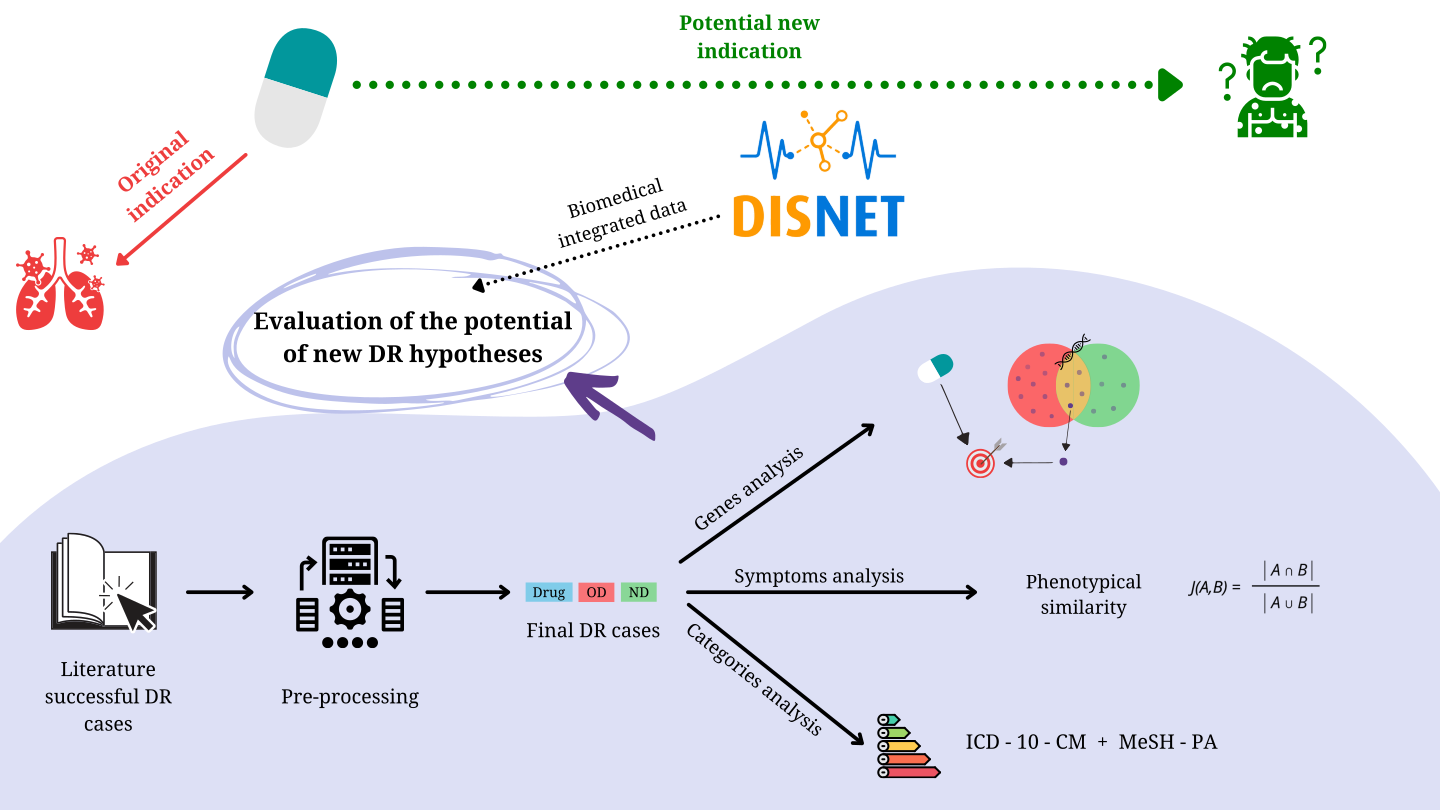
We can draw the following conclussions from our study:
- The analysis of genes, symptoms and categories can provide hints about which DR cases should be prioritized or given more attention to.
- The gene analysis has confirmed that the diseases participating in DR processes (the original disease indicated for a drug, and the new disease that this given drug is repurposed for) present higher association values with target genes for a given drug than the rest of DISNET disease-gene associations.
- The symptom analysis has demonstrated that diseases involved in a DR case are phenotypically more similar (in terms of shared symptoms) that other DISNET diseases.
- The category analysis has suggested that the classes of repositioned drugs and DR diseases frequently follow some specific patterns.
To summarize, the main conclusion of this work was that DISNET’s integrated biomedical data could be considered and used to suggest novel potential repurposing cases. We mainly stated this conclusion because:
- Actual well-known DR cases show significant differences with other non-DR cases regarding their gene and symptom similarities
- DISNET provides accurate DR-related information relevant to the repurposing process (e.g., the drug target gene related to DR-involved diseases).
This work has been published in the Computational and Structural Biotechnology Journal, titled "A data-driven methodology towards evaluating the potential of drug repurposing hypotheses"
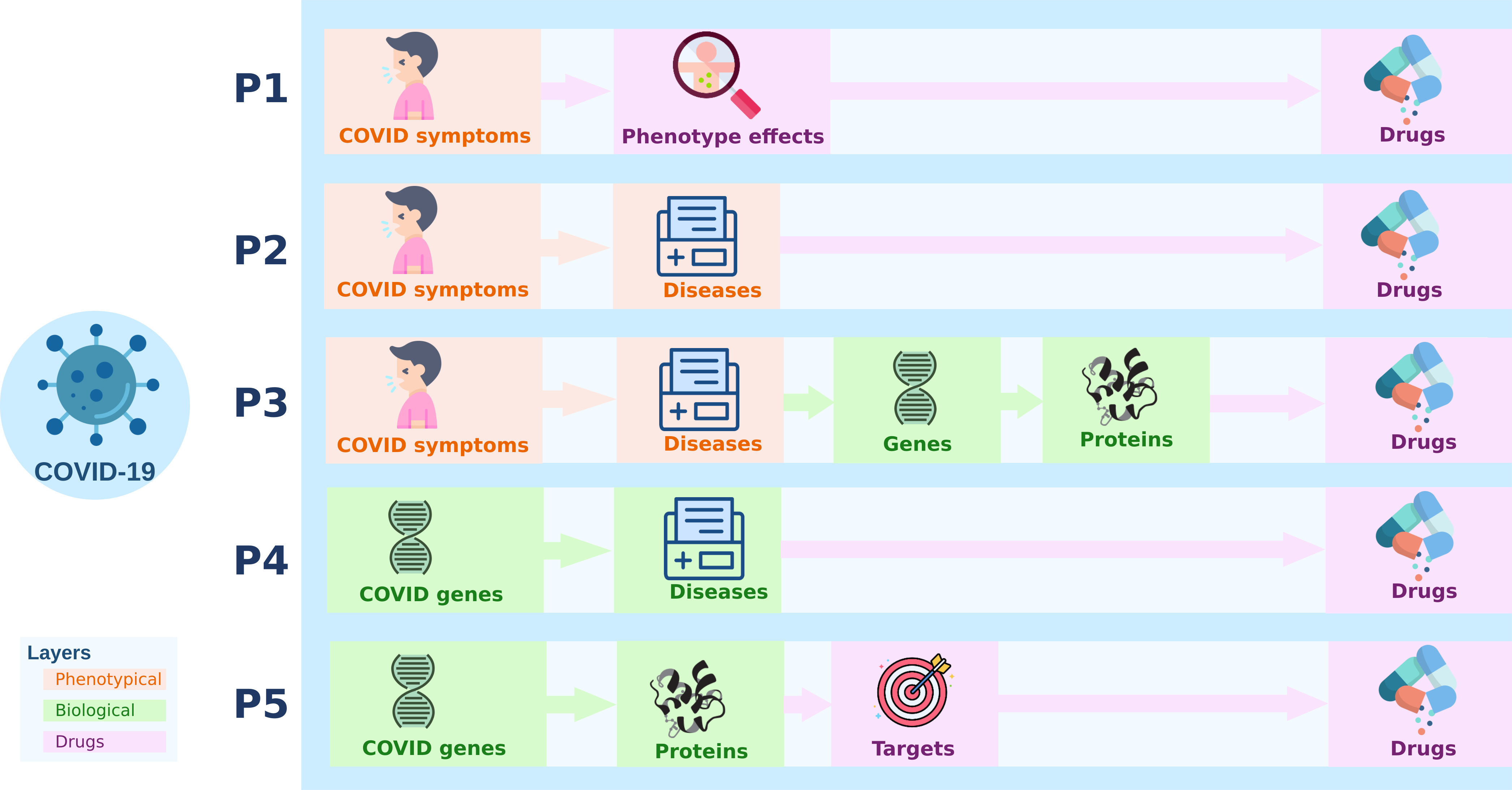
COVID-19
- Drugs used to treat COVID symptoms.
- Drugs used to treat diseases that were symptomatologically similar to COVID-19.
- Drugs that targeted proteins associated with diseases symptomatologically similar to COVID-19.
- Drugs used for diseases that were associated with genes involved in a SARS-like infection.
- Drugs that targeted proteins involved in a SARS-like infection.